“Rewriting” the Genome
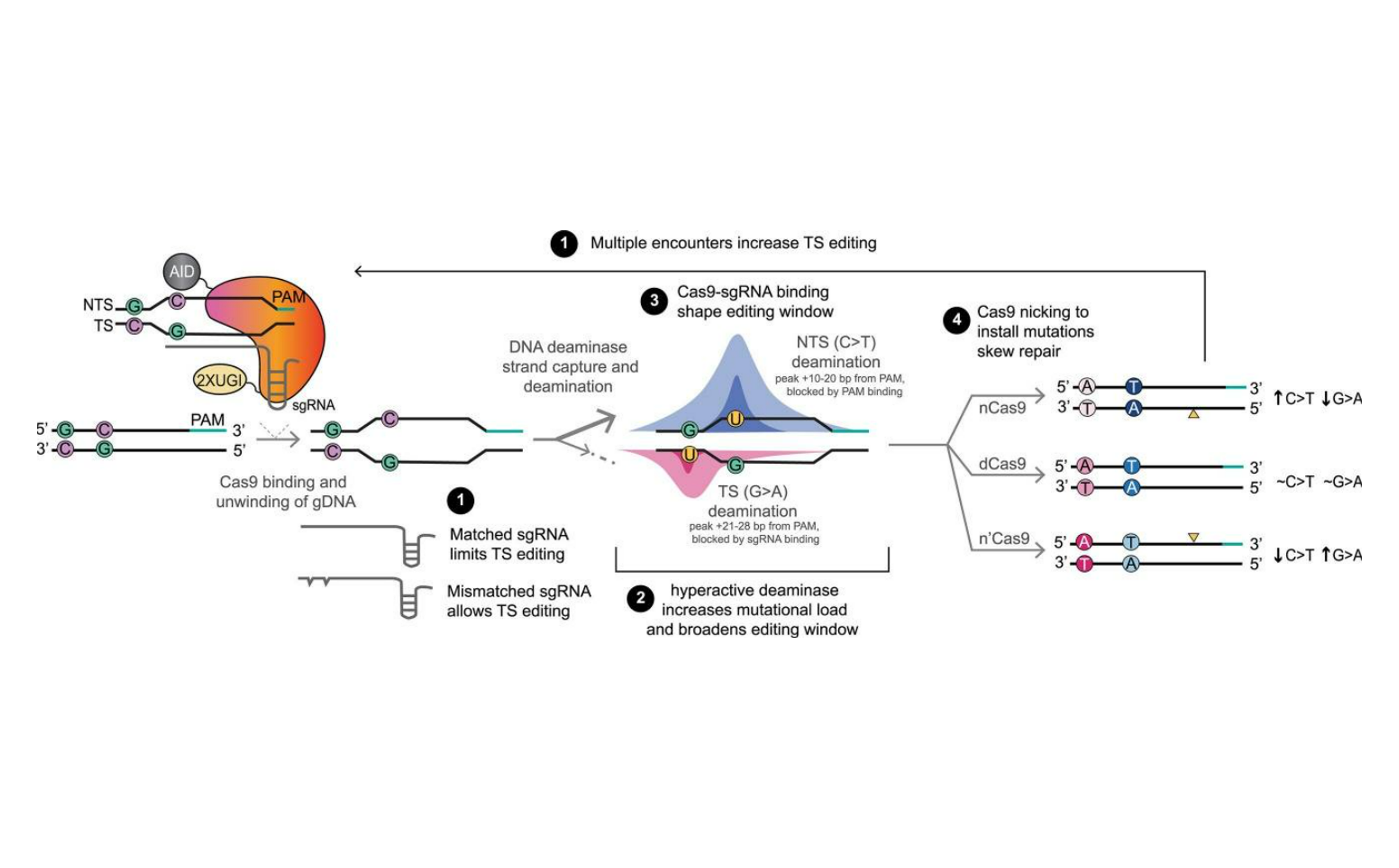
Just as deciphering DNA deaminase and MTase mechanisms allowed us to advance new methods to ‘read’ the epigenome, our mechanistic insights have also helped push frontiers in genome engineering. Base editors (BEs) are powerful examples of harnessing nature’s toolbox. BEs involve the partnership of CRISPR-Cas proteins, which localize the complex via their ‘GPS’ function, and DNA deaminases, which catalyze transition mutations at the targeted locations (C/G-to-T/A or A/T-to-G/C). Despite the remarkable promise of BEs, these tools pose two key challenges, specifically in controlling ‘where’ and more so ‘when’ the genome is edited. In clinical trials and other applications, the only mechanism to control mutational timing is through transient exposure to BEs, which can limit the ability to truly engineer genomes when it would be most biologically relevant.
To address these limitations, we deciphered sites that allow for splitting DNA deaminases into two inactive components, whereby activity could be reconstituted under the control of a small-molecule protein dimerizer (Nat Chem Biol, 2021). We moved these controllable DNA deaminases into the scaffold of BEs and showed notable improvements in ‘where’ editing occurs, with decreased off-target activity using our split-engineered base editors (seBE). Critically, seBEs also allowed for the BE complex to lie dormant in a cell until editing is desired, offering a solution to the ‘when’ challenge. We have also aimed to understand the mechanisms of BEs (NAR, 2024), and, more specifically, how diversifying base editors could not just target single bases, but introduce targeted mutations in genes of interest to allow for protein evolution. Our mechanistic work and the addition of temporal control is already ushering in new insights. For example, in ongoing collaborations, we have introduced seBEs into cells where standard BEs are not tolerated due to toxicity, thus allowing for novel in vivo screens to define anti-cancer targets. In parallel, engineering genome editors with cell-penetrating peptides is an ongoing focus of our collaborative work with the lab of Junwei Shi (Nat Biotechnol, 2024), an advance that allows engineering of hard-to-edit patient-derived primary cells without the toxicities associated with viral delivery or electroporation.
With ongoing support from NIGMS, NIAID, and other sources, the vision of this part of our research program is to help build the genome editing toolbox that can allow for any genomic edit to be introduced into any cell, including the goal of recapitulating the process of antibody somatic hypermutation in target genes of choice. Further, we hope to meld our expertise in DNA methylation and demethylation with our background in genome editing to develop new tools for targeted epigenome editing in cells.